DNA Quantization and Ploidy Analysis
Ploidy analysis is a test performed on cells using either an optical microscope hooked to a computer or a flow cytometer. It is based on the fact that tumors with a diploid pattern of DNA amounts tend to be benign. On the other hand, tumors with highly variable DNA patterns tend be more malignant.
The Ploidics™ product is a stand-alone, software product that performs image processing and image analysis on digital images captured with an optical microscope and stored on a computer. It also processes virtual slide images of any size and depth. Ploidics is an automatic tool to measure abnormal DNA content in individual cells in a mixed population of cells. It includes a fully automatic, powerful, and innovative segmentation process, an editing tool to manually annotate the segmentation result if desired, and an automatic computation of a histogram showing the DNA mass distribution of the cell population.
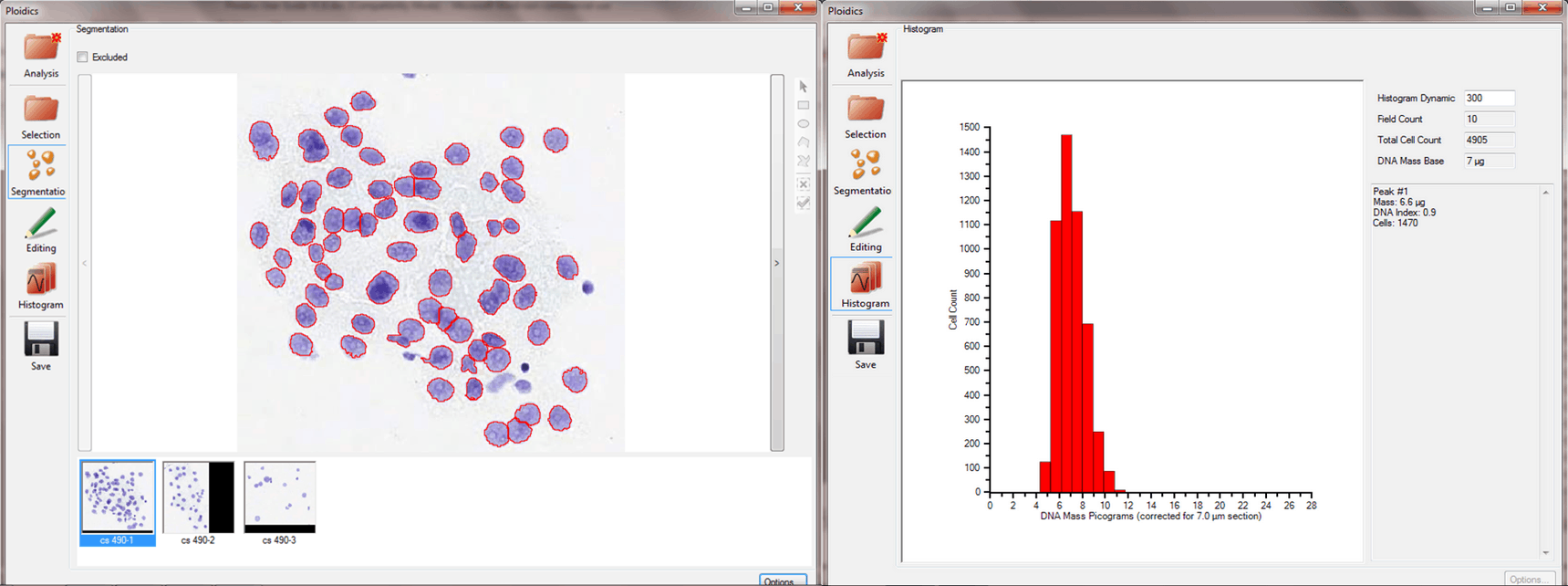
Segmentation results for one image of an image sequence, with Image Gallery shown (left); Typical ploidy histogram (right)
Capabilities
The Ploidics software product has the following capabilities:
- Automatic splitting of touching nuclei
- User selection of nuclei size used in the analysis
- User decision to keep or discard heterogeneous nuclei
- User specification of color band to be processed based on the stain selected
- User modification of segmentation results to select/deselect cells used in the Ploidy analysis
Technical Specifications
Ploidics is a stand-alone, software product that can be installed on any personal computer running Windows® XP or higher. Images need to be accessible from a hard drive or from network storage, and organized in folders to be processed by the software. All Ploidics measurements are calibrated and output in real-world units. The calibration value is manually entered in the Ploidics Software as a single value.
